|
Lea Bogensperger I am a postdoctoral researcher in the Krauthammer Lab at the Department of Quantitative Biomedicine at the University of Zurich. Prior to that, I worked as a doctoral researcher at the Institute of Computer Graphics and Vision, Graz University of Technology. I defended my PhD thesis "Variational Methods in Imaging Meet Machine Learning" in June 2024, which was supervised by Prof. Thomas Pock and with Prof. Carola-Bibiane Schönlieb as my external referee. During my PhD I worked on inverse problems in medical imaging, bilevel optimization as well as generative modeling using diffusion models and flow matching for image segmentation. |

|
ResearchI'm interested in protein design, particularly in optimizing protein fitness using generative modeling and protein language models. Additionally, I have a strong interest in inverse problems in medical imaging. * indicates equal contribution. |

|
Restora-Flow: Mask-Guided Image Restoration with Flow Matching
Arnela Hadzic, Franz Thaler, Lea Bogensperger, Simon Johannes Joham, Martin Urschler Accepted for IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), 2026 arXiv code We introduce RestoraFlow, a training-free method for masked-based inverse problems that guides flow matching sampling by a degradation mask and incorporates a trajectory correction mechanism to enforce consistency with degraded inputs. |
|
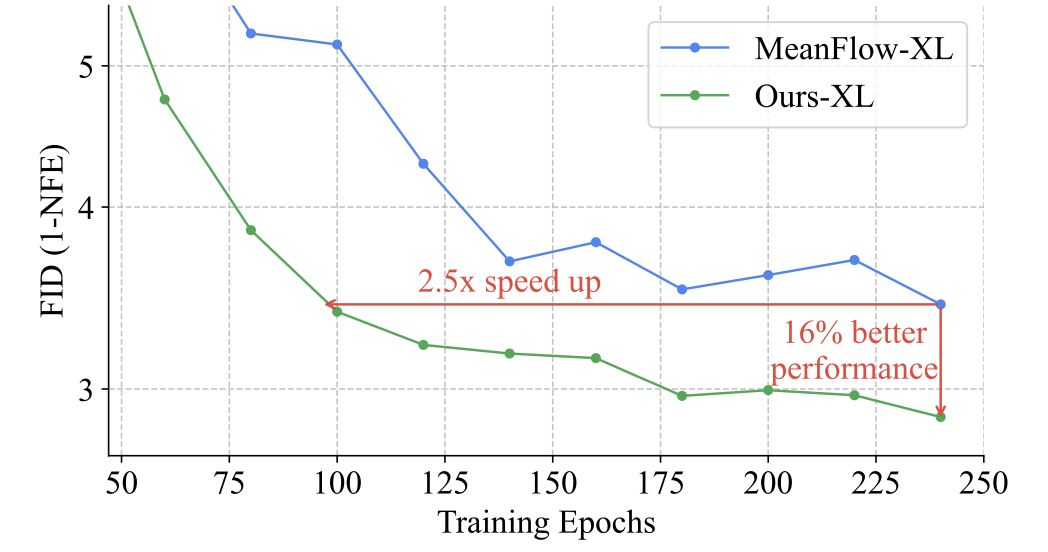
Understanding, Accelerating, and Improving MeanFlow Training
Jin-Young Kim, Hyojun Go, Lea Bogensperger, Julius Erbach, Nikolai Kalischek, Federico Tombari, Konrad Schindler, Dominik Narnhofer arXiv preprint , 2025 arXiv We propose an improved MeanFlow training strategy that rapidly stabilizes instantaneous velocity before progressively emphasizing long-interval averages, enabling faster convergence and higher-quality one-step generation. |
|
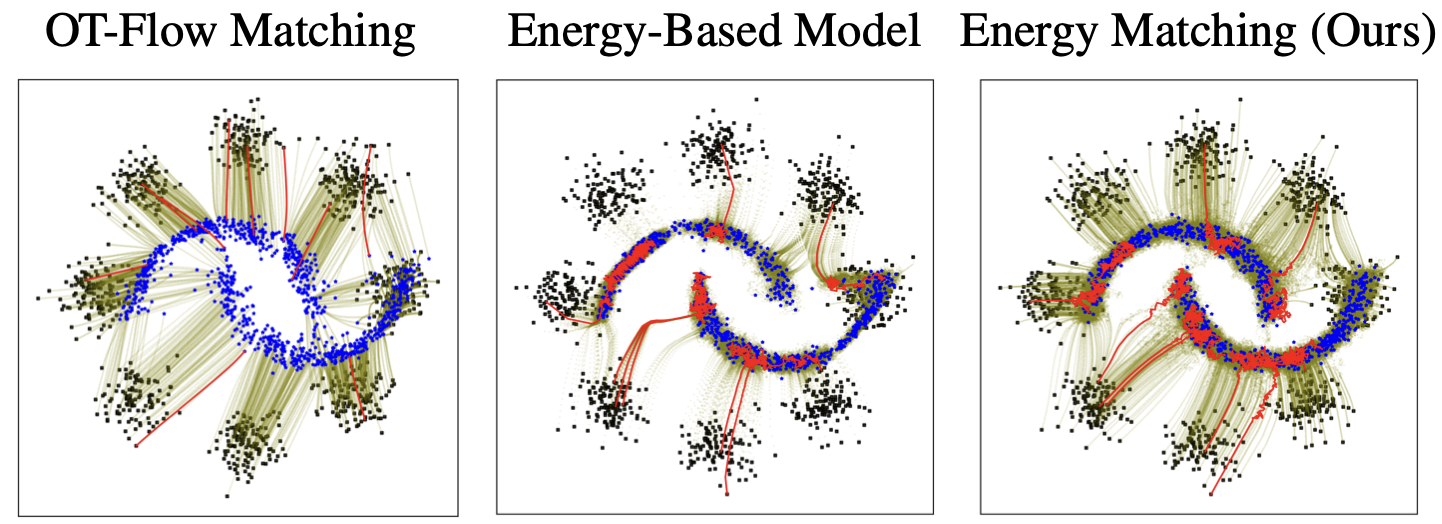
Energy Matching: Unifying Flow Matching and Energy-Based Models for Generative Modeling
Michal Balcerak, Tamaz Amiranashvili, Antonio Terpin, Suprosanna Shit, Lea Bogensperger, Sebastian Kaltenbach, Petros Koumoutsakos, Bjoern Menze Conference on Neural Information Processing Systems (NeurIPS), 2025 arXiv code An energy matching framework is introduced, combining optimal transport paths far from the data manifold with an entropic energy term to explicitly capture data likelihood, enabling flexible priors and high-fidelity generation without auxiliary networks. |
|
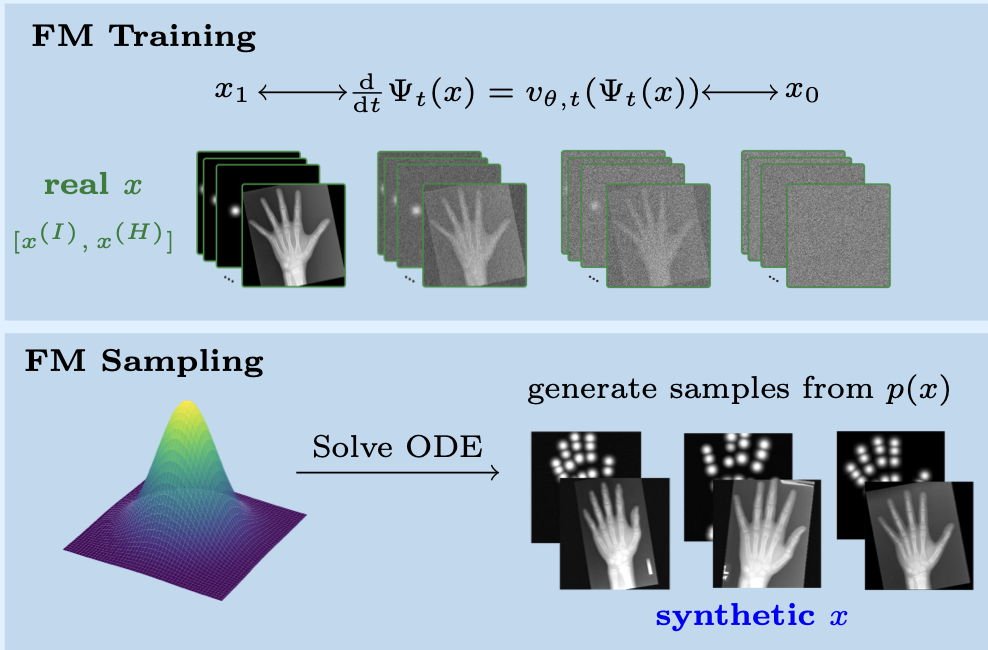
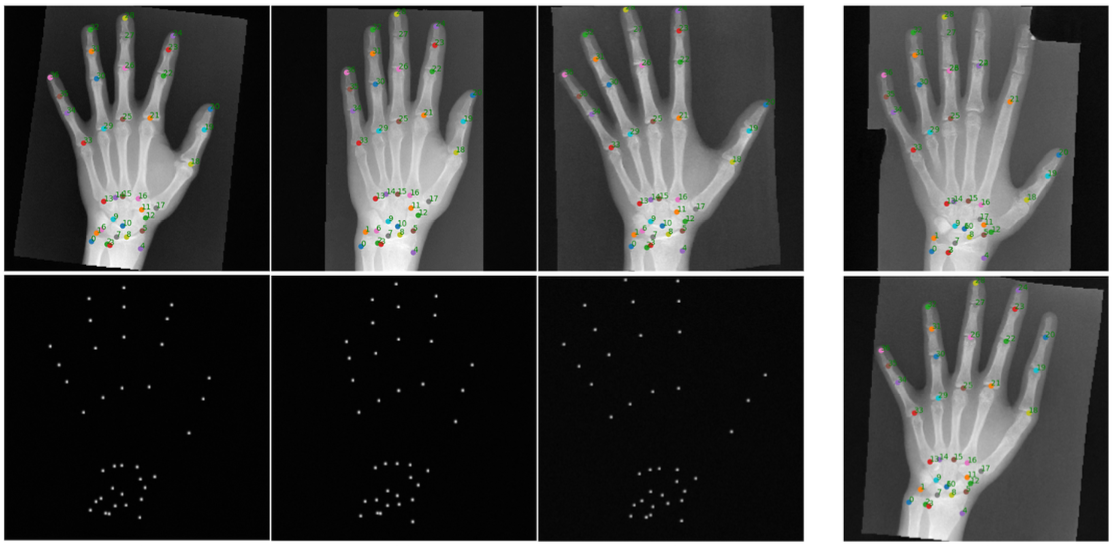
Flow Matching-Based Data Synthesis for Robust Anatomical Landmark Localization
Arnela Hadzic, Lea Bogensperger, Andrea Berghold, Martin Urschler IEEE Journal of Biomedical and Health Informatics, 2025 paper We introduce a multi-channel generative approach based on Flow Matching to synthesize medical images paired with heatmaps, enabling robust data augmentation and enhanced generalization for anatomical landmark localization - especially in settings with limited training data or occlusions. |
|
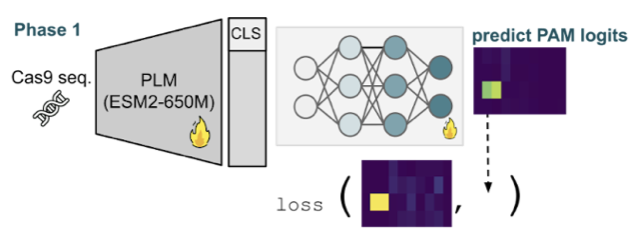
Uncovering Cas9 PAM diversity through metagenomic mining and machine learning
Tao Fang*, Lea Bogensperger*, Lilith Feer, Ahmed Allam, Valentyn Bezshapkin, Zsolt Balázs, Christian von Mering, Shinichi Sunagawa, Michael Krauthammer, Gerald Schwank under review, 2025 bioRxiv code CRISPR-PAMdb, a large-scale database of Cas9 proteins and PAM profiles, is introduced alongside CICERO, a protein language model–based predictor that enables accurate PAM preference inference and broad exploration of PAM diversity for genome editing. |
|
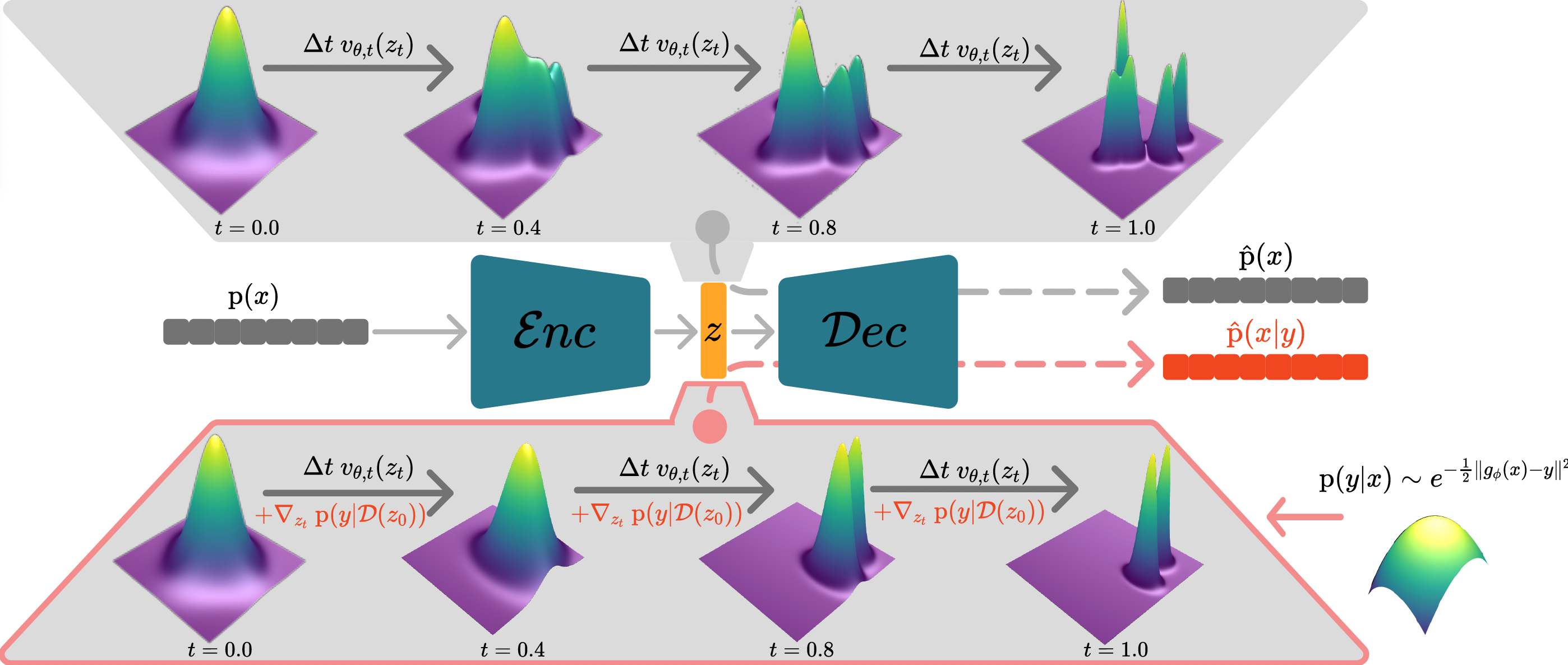
A Variational Perspective on Generative Protein Fitness Optimization
Lea Bogensperger, Dominik Narnhofer, Ahmed Allam, Konrad Schindler, Michael Krauthammer International Conference on Machine Learning (ICML), 2025 arXiv paper code A variational framework for generative protein fitness optimization using a flow matching prior and a classifier-guidance model in a continuous latent space is introduced. |
|
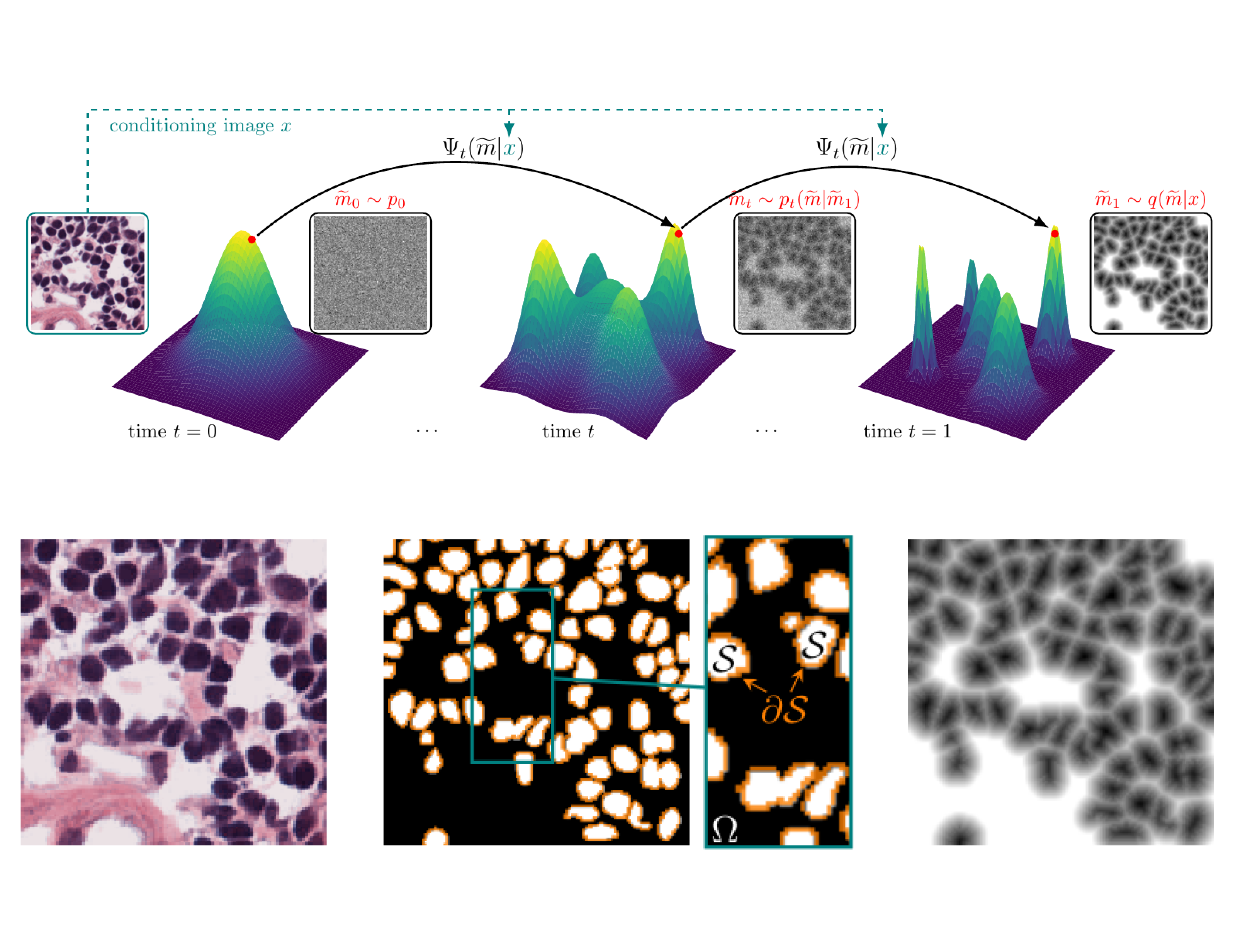
FlowSDF: Flow Matching for Medical Image Segmentation Using Distance Transforms
Lea Bogensperger*, Dominik Narnhofer*, Alexander Falk, Konrad Schindler, Thomas Pock International Journal of Computer Vision, 2025 arXiv paper code A flow matching framework for generative medical image segmentation using signed distance functions that enables canonical smoothing of SDF mask distributions through noise injection. |
|
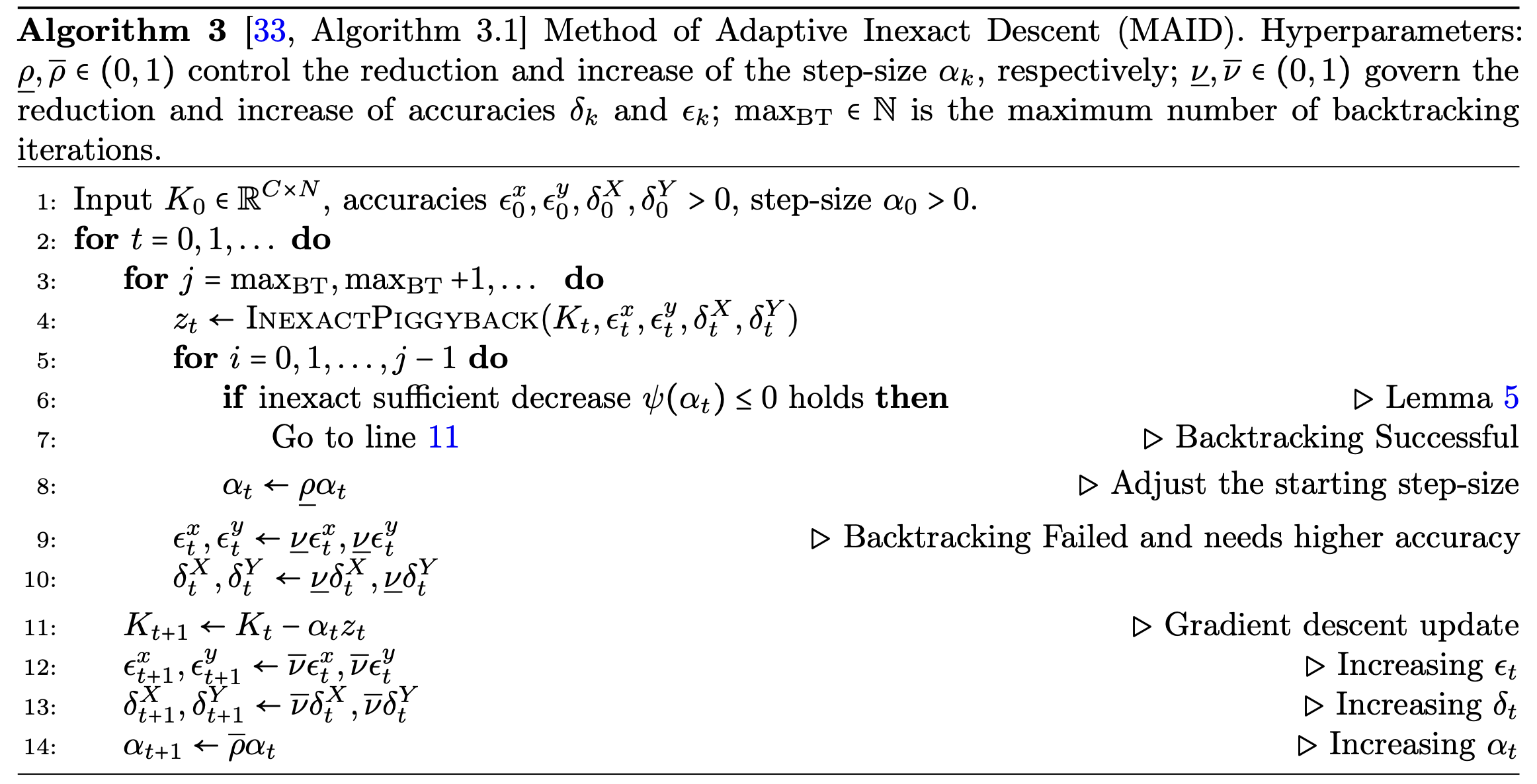
An Adaptively Inexact Method for Bilevel Learning Using Primal-Dual Style Differentiation
Lea Bogensperger, Matthias J. Ehrhardt, Thomas Pock, Mohammad Sadegh Salehi, Hok Shing Wong Journal of Mathematical Imaging and Vision (JMIV), 2025 arXiv paper We introduce a bilevel learning framework for variational image reconstruction that uses a primal-dual approach with a-posteriori error bounds and adaptive step sizes. |
|
Synthetic Augmentation for Anatomical Landmark Localization using DDPMs
Arnela Hadzic, Lea Bogensperger, Simon Johannes Joham, Martin Urschler Simulation and Synthesis in Medical Imaging (SASHIMI), 2024 arXiv paper A DDPM-based framework for generating medical images with landmark heatmaps, using a Markov Random Field for matching and a Statistical Shape Model for plausibility checks. |
|
Score-Based Generative Models for Medical Image Segmentation using Signed Distance Functions
Lea Bogensperger, Dominik Narnhofer, Filip Ilic, Thomas Pock DAGM German Conference on Pattern Recognition, 2023 (Honorable Mention) arXiv paper code A score-based generative model for binary medical image segmentation using signed distance functions, where diffusion corrupts SDF masks instead of binary ones for more natural distortions. |
|
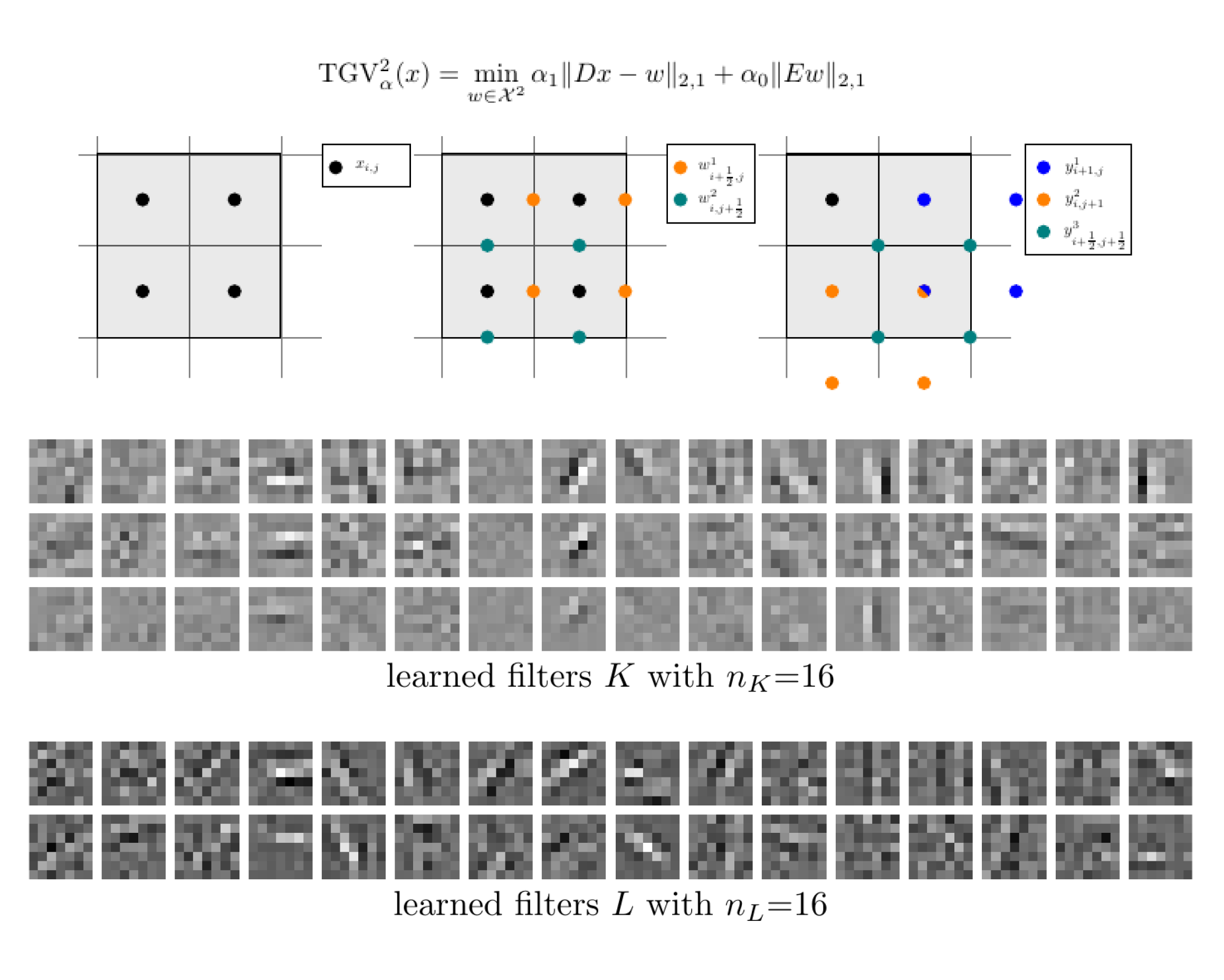
Learned Discretization Schemes for the Second-Order Total Generalized Variation
Lea Bogensperger, Antonin Chambolle, Alexander Effland, Thomas Pock International Conference on Scale Space and Variationa Methods in Computer Vision, 2023 arXiv paper code We introduce a framework for incorporating general discretizations of second-order TGV with variational consistency, and learn interpolation filters via a piggyback algorithm. |
|
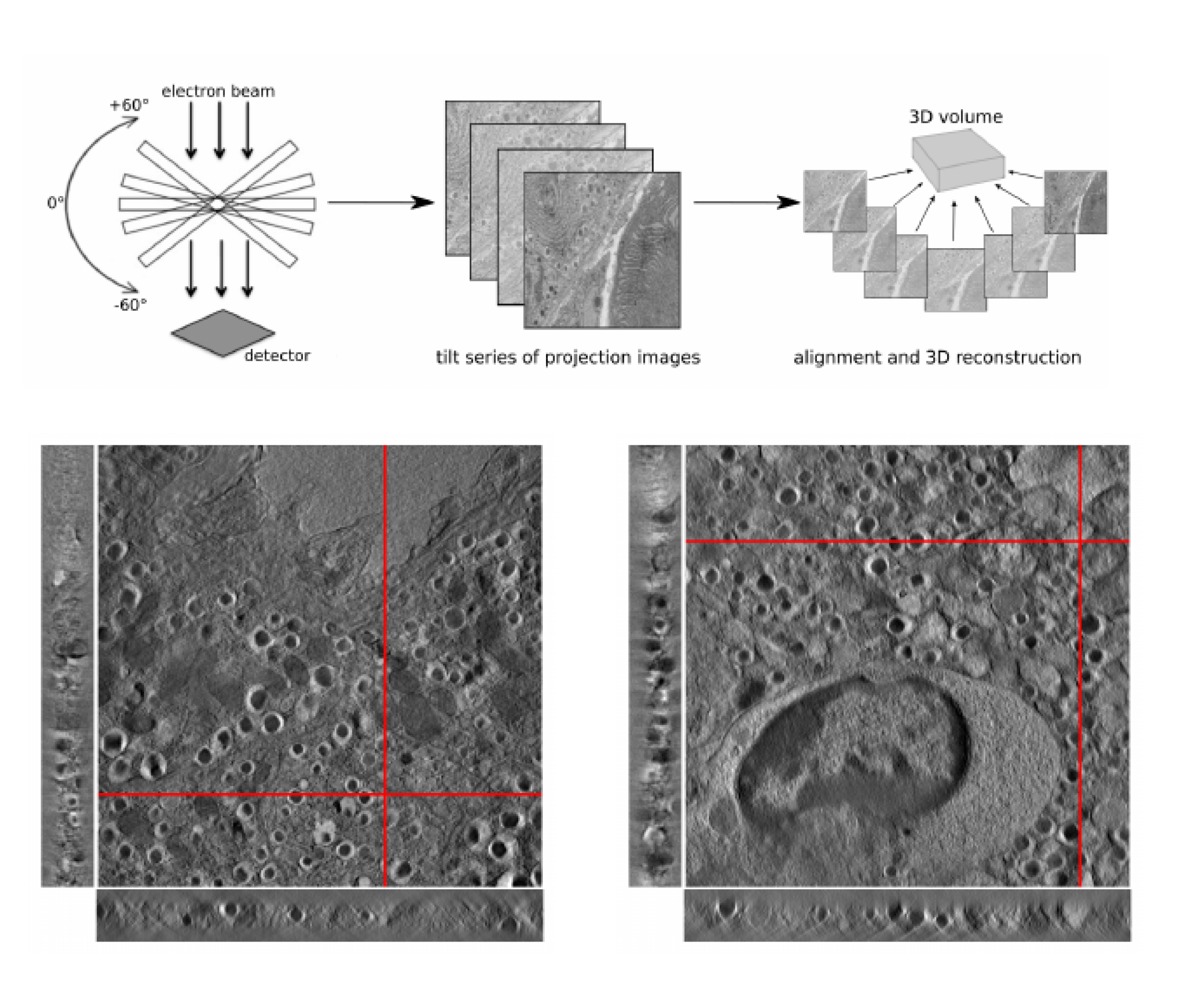
A joint alignment and reconstruction algorithm for electron tomography to visualize in-depth cell-to-cell interactions
Lea Bogensperger, Erich Kobler, Dominique Pernitsch, Petra Kotzbeck, Thomas R. Pieber, Thomas Pock, Dagmar Kolb Histochemistry and Cell Biology, 2022 paper code We develop a joint alignment and reconstruction algorithm for electron tomography without fiducial markers, applied to studying immune–beta cell interactions in NOD mice for type 1 diabetes research. |
|
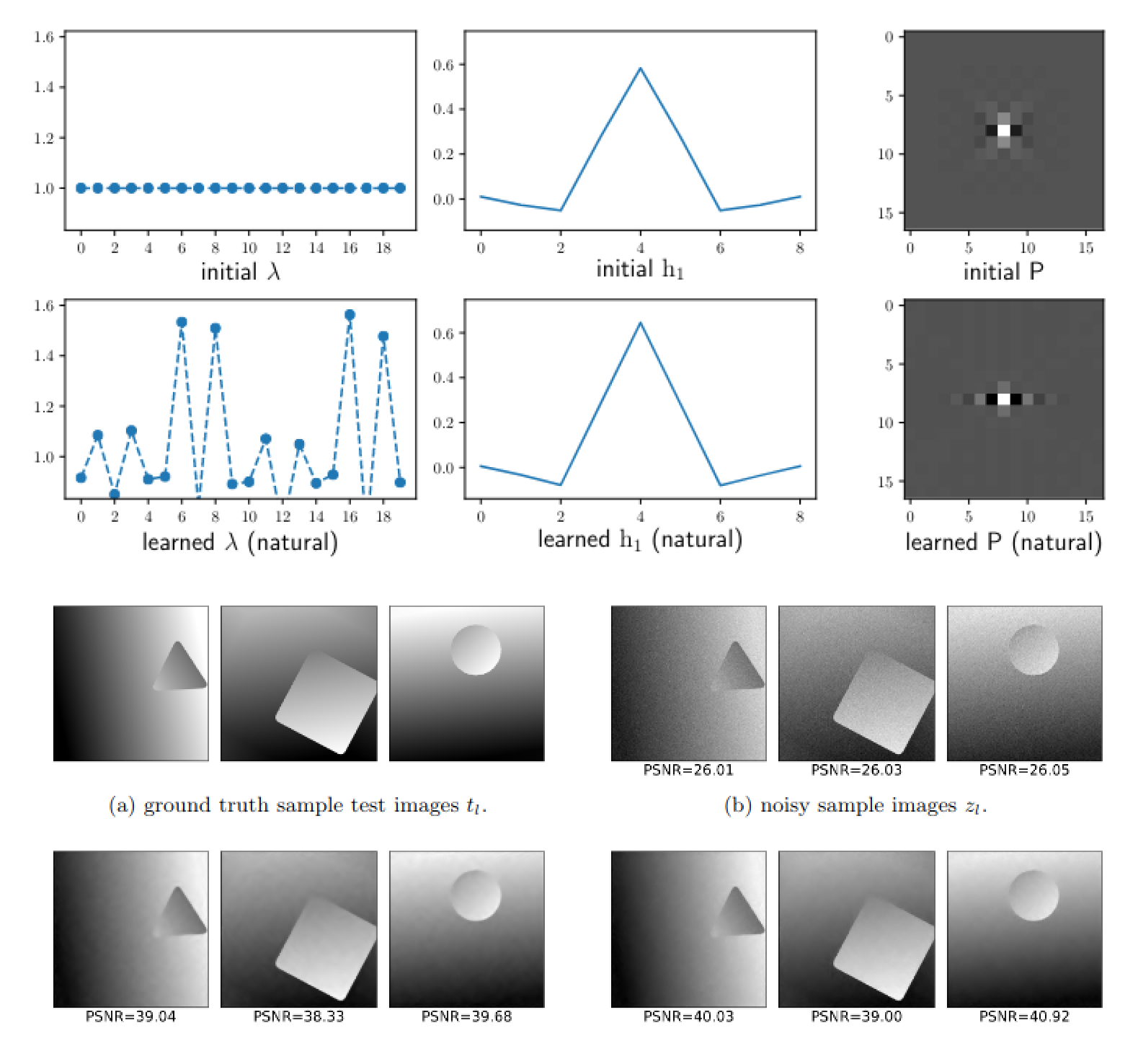
Convergence of a Piggyback-style method for the differentiation of solutions of standard saddle-point problems
Lea Bogensperger, Antonin Chambolle, Thomas Pock SIAM Journal on Mathematics of Data Science, 2022 paper preprint We analyze a piggyback-style method for differentiating saddle-point problems, and apply it to learning optimized shearlet transforms for imaging. |
|
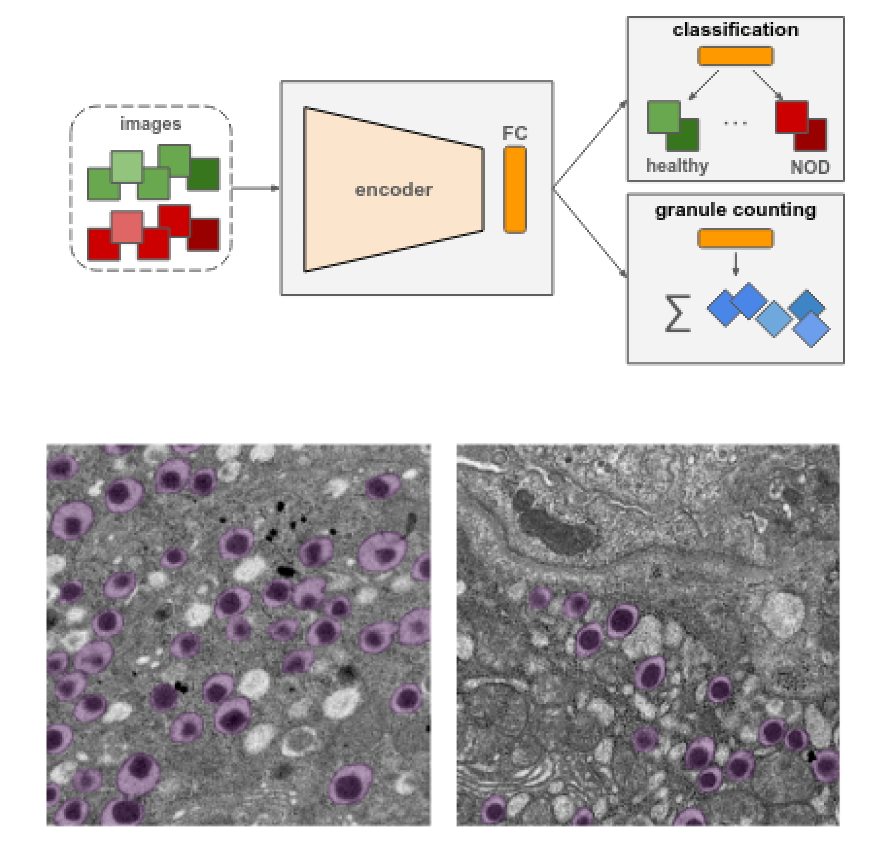
On the Influence of Beta Cell Granule Counting
for Classification in Type 1 Diabetes
Lea Bogensperger, Marc Masana, Filip Ilic, Dagmar Kolb, Thomas R. Pieber, Thomas Pock Computer Vision and Pattern Analysis Across Domains, 2021 paper We use deep learning to study insulin granules in NOD mouse beta cells, where a multi-task regression aids in distinguishing healthy from diabetic samples. |
|
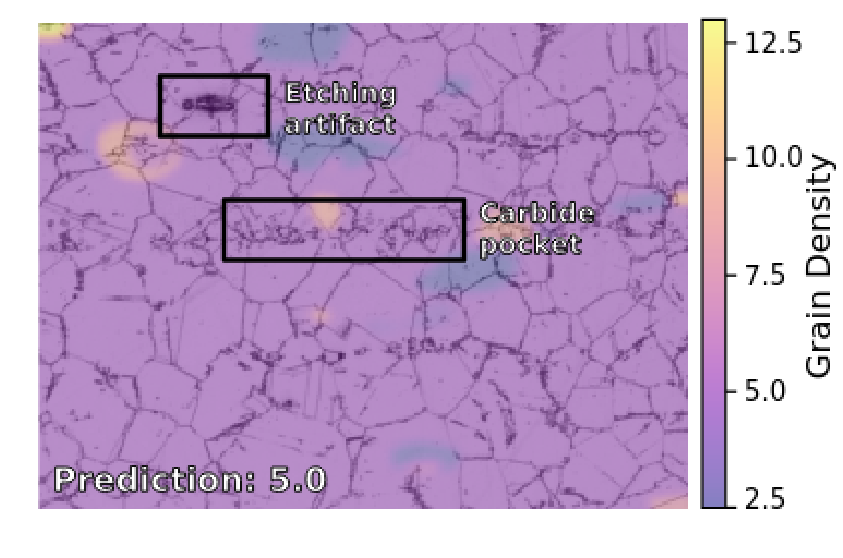
A study on robust feature representations for grain density estimates in austenitic steel
Filip Ilic, Marc Masana, Lea Bogensperger, Harald Ganster, Thomas Pock Computer Vision and Pattern Analysis Across Domains, 2021 paper code We show that deep neural networks can estimate grain density in austenitic steel, with classification and regression models learning distinct feature representations. |
Teaching |
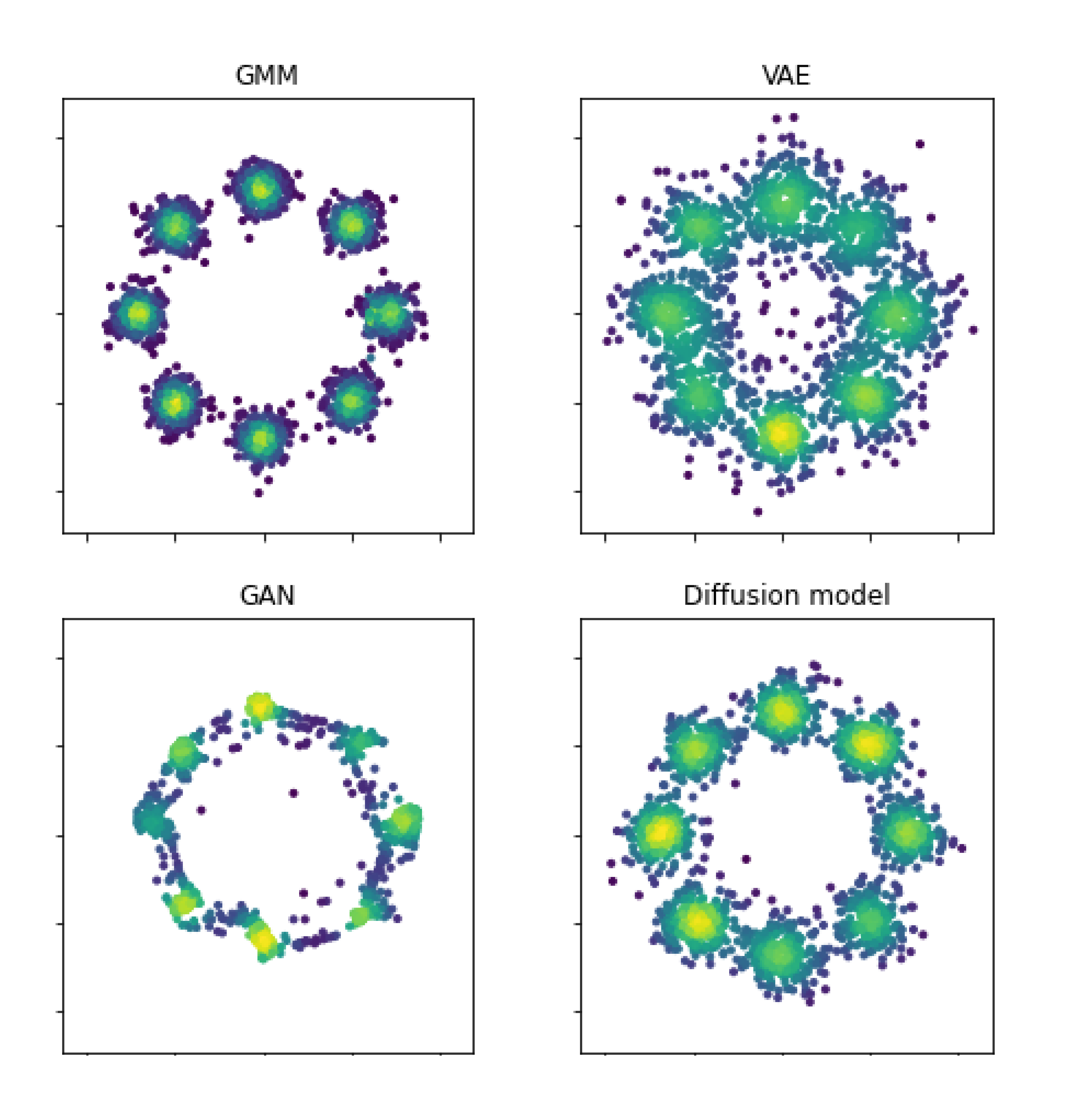 |
Lecture Notes for Generative Modeling
code |